| dyads_test_vs_ctrl_m1_shift9 (dyads_test_vs_ctrl_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; dyads_test_vs_ctrl_m1; m=0 (reference); ncol1=12; shift=9; ncol=23; ---------trACACGTGTya--
; Alignment reference
a 0 0 0 0 0 0 0 0 0 85 89 278 12 319 20 20 10 17 19 81 121 0 0
c 0 0 0 0 0 0 0 0 0 66 76 31 308 4 297 5 9 5 14 96 70 0 0
g 0 0 0 0 0 0 0 0 0 70 96 14 5 9 5 297 4 308 31 76 66 0 0
t 0 0 0 0 0 0 0 0 0 121 81 19 17 10 20 20 319 12 278 89 85 0 0
|
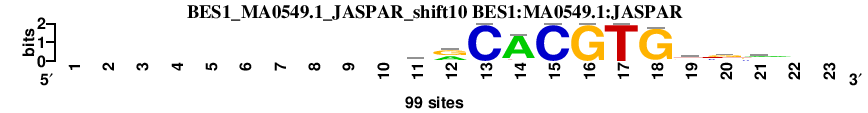
| BES1_MA0549.1_JASPAR_shift10 (BES1:MA0549.1:JASPAR) |
 |
0.893 |
0.818 |
6.810 |
0.936 |
0.955 |
24 |
2 |
27 |
20 |
20 |
18.600 |
10 |
; dyads_test_vs_ctrl_m1 versus BES1_MA0549.1_JASPAR (BES1:MA0549.1:JASPAR); m=10/110; ncol2=11; w=11; offset=1; strand=D; shift=10; score= 18.6; ----------crCACGTGksv--
; cor=0.893; Ncor=0.818; logoDP=6.810; NsEucl=0.936; NSW=0.955; rcor=24; rNcor=2; rlogoDP=27; rNsEucl=20; rNSW=20; rank_mean=18.600; match_rank=10
a 0 0 0 0 0 0 0 0 0 0 23 41 0 89 0 0 0 3 16 17 36 0 0
c 0 0 0 0 0 0 0 0 0 0 45 8 99 0 99 0 0 0 5 33 29 0 0
g 0 0 0 0 0 0 0 0 0 0 10 50 0 4 0 99 0 96 36 46 33 0 0
t 0 0 0 0 0 0 0 0 0 0 21 0 0 6 0 0 99 0 42 3 1 0 0
|
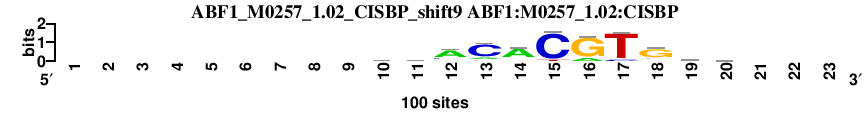
| ABF1_M0257_1.02_CISBP_shift9 (ABF1:M0257_1.02:CISBP) |
 |
0.917 |
0.841 |
3.594 |
0.946 |
0.967 |
11 |
1 |
74 |
10 |
10 |
21.200 |
11 |
; dyads_test_vs_ctrl_m1 versus ABF1_M0257_1.02_CISBP (ABF1:M0257_1.02:CISBP); m=11/110; ncol2=11; w=11; offset=0; strand=D; shift=9; score= 21.2; ---------gdACACGTGkb---
; cor=0.917; Ncor=0.841; logoDP=3.594; NsEucl=0.946; NSW=0.967; rcor=11; rNcor=1; rlogoDP=74; rNsEucl=10; rNSW=10; rank_mean=21.200; match_rank=11
a 0 0 0 0 0 0 0 0 0 21 26 68 16 73 0 18 0 12 16 18 0 0 0
c 0 0 0 0 0 0 0 0 0 21 16 18 75 8 91 0 10 4 19 28 0 0 0
g 0 0 0 0 0 0 0 0 0 37 33 7 0 10 0 82 0 71 40 28 0 0 0
t 0 0 0 0 0 0 0 0 0 21 25 7 9 9 9 0 90 13 25 26 0 0 0
|
| LOC_Os02g49560_M0327_1.02_CISBP_shift9 (LOC_Os02g49560:M0327_1.02:CISBP) |
 |
0.881 |
0.807 |
6.303 |
0.935 |
0.954 |
32 |
4 |
38 |
22 |
21 |
23.400 |
13 |
; dyads_test_vs_ctrl_m1 versus LOC_Os02g49560_M0327_1.02_CISBP (LOC_Os02g49560:M0327_1.02:CISBP); m=13/110; ncol2=11; w=11; offset=0; strand=D; shift=9; score= 23.4; ---------tkcCACGTGwy---
; cor=0.881; Ncor=0.807; logoDP=6.303; NsEucl=0.935; NSW=0.954; rcor=32; rNcor=4; rlogoDP=38; rNsEucl=22; rNSW=21; rank_mean=23.400; match_rank=13
a 0 0 0 0 0 0 0 0 0 22 18 20 0 100 0 0 0 2 27 21 0 0 0
c 0 0 0 0 0 0 0 0 0 22 18 64 93 0 100 0 0 22 7 29 0 0 0
g 0 0 0 0 0 0 0 0 0 23 39 4 7 0 0 100 0 73 19 21 0 0 0
t 0 0 0 0 0 0 0 0 0 33 25 12 0 0 0 0 100 3 47 29 0 0 0
|
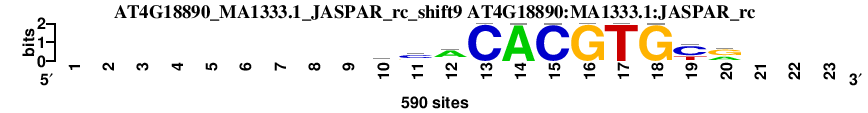
| AT4G18890_MA1333.1_JASPAR_rc_shift9 (AT4G18890:MA1333.1:JASPAR_rc) |
 |
0.884 |
0.811 |
0.339 |
0.930 |
0.946 |
28 |
3 |
101 |
28 |
31 |
38.200 |
26 |
; dyads_test_vs_ctrl_m1 versus AT4G18890_MA1333.1_JASPAR_rc (AT4G18890:MA1333.1:JASPAR_rc); m=26/110; ncol2=11; w=11; offset=0; strand=R; shift=9; score= 38.2; ---------ycACACGTGyr---
; cor=0.884; Ncor=0.811; logoDP=0.339; NsEucl=0.930; NSW=0.946; rcor=28; rNcor=3; rlogoDP=101; rNsEucl=28; rNSW=31; rank_mean=38.200; match_rank=26
a 0 0 0 0 0 0 0 0 0 81 52 405 0 589 0 0 0 1 0 225 0 0 0
c 0 0 0 0 0 0 0 0 0 179 338 60 590 0 589 0 0 0 383 43 0 0 0
g 0 0 0 0 0 0 0 0 0 91 140 74 0 0 0 590 0 589 28 319 0 0 0
t 0 0 0 0 0 0 0 0 0 239 60 51 0 1 1 0 590 0 179 3 0 0 0
|




